Display a distribution graph.
Syntax
distplot(options) o1 [o2 o3 ... ]
object_name.distplot(options) analytical_spec(arg) [categorical_spec(arg)]
where o1, o2, ..., are series or group objects.
When used as a command, distplot only allows you to display the default histogram view.
When used as an object view, you must specify the type of distribution graph you wish to create in the
analytical_spec. You may select from: histogram, histogram polygon, histogram edge polygon, average shifted histogram, kernel density, theoretical distribution, empirical CDF, empirical survivor, empirical log survivor, or empirical quantile (see
“Analytical Spec”).
The optional
categorical_spec allows you to specify a categorical graph (see
“Categorical Spec”)
Options
Multiple series options
s | Plot in a single graph. (Categorical graph settings will override this option.) |
Template and printing options
o=template | Use appearance options from the specified template. template may be a predefined template keyword (“default” - current global defaults, “classic”, “modern”, “reverse”, “midnight”, “spartan”, “monochrome”) or a graph in the workfile. |
t=graph_name | Use appearance options and copy text and shading from the specified graph. |
b / -b | [Apply / Remove] bold modifiers of the base template style specified using the “o=” option above. |
w / -w | [Apply / Remove] wide modifiers of the base template style specified using the “o=” option above. |
reset | Resets all graph options to the global defaults. May be used to remove existing customization of the graph. |
p | Print the graph. |
The options which support the “–” may be preceded by a “+” or “–” indicating whether to turn on or off the option. The “+” is optional.
Panel options
The following option applies when graphing panel structured data.
panel=arg (default taken from global settings) | Panel data display: “stack” (stack the cross-sections), “individual” or “i” (separate graph for each cross-section). (Note: more general versions of these panel graphs may be constructed as categorical graphs.) |
Analytical Spec
Specify the distribution graph you wish to create in the analytical spec. For a description of distribution graphs, see
“Analytical Graph Types”. The analytical spec contains components of the form:
dist_type(dist_options)
where dist_type may be one of the following keywords:
hist | Histogram. |
freqpoly | Histogram Polygon. |
edgefreqpoly | Histogram Edge Polygon. |
ash | Average Shifted Histogram. |
kernel | Kernel Density |
theory | Theoretical Distribution. |
cdf | Empirical cumulative distribution function. |
survivor | Empirical survivor function. |
logsurvivor | Empirical log survivor function. |
quantile | Empirical quantile function. |
hist, freqpoly, edgefreqpoly, ash, kernel, and theory graphs may be combined in a single graph frame by providing multiple components.
Each distribution type has its own set of options, to be entered in dist_options:
Histogram, Histogram Polygon, Histogram Edge Polygon, and Avg. Shifted Histogram Options
scale=arg | arg specifies the scaling size, and may be “dens”, “freq”, or “relfreq”. (Note that the scaling setting is overridden if the histogram is displayed alongside a density, e.g., kernel density or theoretical distribution, plot.) |
binw=arg | arg specifies the bin width, and may be “eviews” (default), “sigma” (normal reference rule with 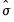 as the measure of dispersion), “iqr” (normal reference rule based on the interquartile range), “silverman” (normal reference rule with Silverman’s robust measure of dispersion), “freedman” (Freedman-Diaconis), “user” (user-specifed). |
binval=arg | arg specifies the numeric value of the bin width, when the option “binw=user” is specified. |
anchor=arg | arg specifies the anchor position. |
rightclosed | Right-closed bin intervals. |
nshifts=int (default=25) | Specifies the number of shift evaluations. (Only applies to average shifted histograms.) |
fill | Fill the graph. (Does not apply to the hist type.) |
nofill | Don’t fill the graph. (Does not apply to the hist type.) |
leg=arg | Specify the legend display settings, where arg can be: “def” - default, “n” - none, “s” - short, “det”- detailed. |
Histogram, Histogram Polygon, Histogram Edge Polygon, and Avg. Shifted Histogram Examples
inf.distplot hist
displays the default histogram view of the frequencies in each bin.
inf.distplot hist(scale=dens, anchor=100, binw=sigma)
constructs a density histogram computed using anchor position 100 and bin width determined by the normal reference rule using
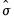
as the measure of dispersion.
group g1 inf unemp
g1.distplot hist(scale=relfreq)
displays a relative frequency histogram for the series in INF and UNEMP, each in their own graph frame, while:
g1.distplot(s) histpoly
displays the two frequency histograms in the same graph frame.
g1.distplot freqpoly(fill)
constructs filled frequency polygons for the series in G1, displayed in individual frames.
inf.distplot edgefreqpoly(leg=detailed)
shows the edge frequency polygon for INF with detailed legend entries.
g1.distplot ash(scale=dens, rightclosed, nshifts=100)
constructs average shifted density histograms using 100 shifts, with right-closed bins.
Kernel Options
k=arg (default=“e”) | Kernel type: “e” (Epanechnikov), “r” (Triangular), “u” (Uniform), “n” (Normal–Gaussian), “b” (Biweight–Quartic), “t” (Triweight), “c” (Cosinus). |
b=number | Specify a number for the bandwidth. |
b | Bracket bandwidth. |
ngrid=integer (default=100) | Number of grid points to evaluate. |
x | Exact evaluation. |
fill | Fill the area. |
nofill | Don’t fill the area. |
leg=arg | Specify the legend display settings, where arg can be: “def” - default, “n” - none, “s” - short, “det”- detailed. |
Kernel Examples
group gg weight height
gg.distplot kernel(ngrid=200, fill)
constructs kernel density estimates of HEIGHT and WEIGHT using 200 grid points and linear binning, and displays filled graphs in individual graph frames.
gg.displot(s) kernel(k=u, x)
computes the estimates using a uniform kernel with exact evaluation at each of the grid points, and displays the graphs in the same frame.
gg.displot kernel(leg=det)
displays the kernel plots along with detailed legend information.
Theory Options
dist=arg | arg can be: “normal”, “exp” - exponential, “logit” - logistic, “uniform” - uniform, “xman” - extreme max, “xmin” - extreme min, “chisq” - chi-squared, “pareto” - Pareto, “weibull” - Weibull, “gamma” - gamma, “tdist” - Student’s t-distribution. |
p1=int | Set first parameter. |
p2=int | Set second parameter. |
p3=int | Set third parameter. |
fill | Fill the area. |
nofill | Don’t fill the area. |
leg=arg | Specify the legend display settings, where arg can be: “def” - default, “n” - none, “s” - short, “det”- detailed. |
m=int | Set the iterations maximum. (Applies to logistic, extreme max, extreme min, chi-squared, Weibull, gamma or t-distributions.) |
c =int | Sets the convergence criterion. (Applies to logistic, extreme max, extreme min, chi-squared, Weibull, gamma or t-distributions.) |
s | Use user-specified starting values supplied in the C coefficient vector in the workfile (default uses EViews supplied starting values). (Applies to logistic, extreme max, extreme min, chi-squared, Weibull, gamma, or t-distributions.) |
Theory Examples
gdp50.distplot theory(leg=det)
displays a normal density plot fitted to the data in GDP50 with detailed legend information.
gdp50.distplot theory(p1=0)
fits a normal density using GDP50, restricting the mean of the distribution to be zero.
group gro1 weight height
gro1.distplot theory(dist=exp, fill)
constructs filled plots of the exponential densities fitted to the data in WEIGHT and HEIGHT, and displays them in separate frames.
gro1.distplot(s) theory(dist=weibull, p1=5, c=1e-5)
fits weibull densities to the data in the series setting the first parameter to 5 and estimating the second with a convergence tolerance of 1e-5. The graphs are displayed in a single frame.
Empirical CDF, Survivor, Log Survivor, and Quantile Options
q=arg | Set the quantile method, where arg can be: “r” - Rankit-Cleveland, “o” - Ordinary, “v” - van der Waerden, “b” - Blom, “t” - Tukey, “g” - Gumbel. |
n or noci | Do not include confidence intervals. |
ci=number (default=0.95) | Set confidence interval levels. |
leg=arg | Specify the legend display settings, where arg can be: “def” - default, “n” - none, “s” - short, “det”- detailed. |
Empirical CDF, Survivor, Log Survivor, and Quantile Examples
gdp50.distplot cdf
shows the cumulative distribution plot for GDP50, along with the default 95% confidence intervals.
gdp50.distplot survivor(noci)
displays the survivor plot for GDP50 without displaying confidence intervals.
group gro1 weight height
gro1.distplot logsurvivor(ci=0.9, leg=det)
displays the log-survivor plots for WEIGHT and HEIGHT along with 90% confidence intervals, and a detailed legend. The plots will be displayed in individual graph frames.
gro1.distplot(s) quantile
shows the quantile plots for WEIGHT and HEIGHT in the same graph frame.
Examples
Basic examples
distplot height weight length
displays default histograms for the three series.
group g1 age height weight length
g1.distplot hist(scale=dens, binw=sigma, leg=short) kernel theory
displays distribution plots for AGE, HEIGHT, WEIGHT, and LENGTH in separate frames, along with a short legend identifying each distribution plot. Each frame contains a histogram constructed using the
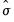
-normal reference rule, a kernel density plot, and a plot of the theoretical normal distribution fitted to the data. (Note that the “scale=dens” option in the
hist specification is redundant since combining a histogram with either the
kernel or
theory plot automatically sets the scaling.)
height.distplot theory theory(dist=weibull)
plots theoretical normal and weibull densities fit to the data in HEIGHT.
height.distplot quantile
displays a plot of the quantiles of height along with the confidence intervals.
g1.displot(s) cdf
plots the empirical CDF of the AGE, HEIGHT, WEIGHT, and LENGTH, and displays them in a single frame.
Panel examples
height.distplot(panel=individual) hist
displays histograms for each cross-section in separate frames while,
weight.distplot kern ash
displays a kernel density graph and average shifted histogram using the panel stacked WEIGHT data.
Categorical spec examples
height.distplot hist across(firm, dispname)
displays a categorical histogram graph of SER1 using distinct values of FIRM to define the categories, and displaying the resulting graphs in multiple frames.
height.distplot hist across(firm, dispname, iscale)
shows the same graph with individual scaling for each of the frames.
weight.distplot kernel ash within(firm, inctot, label=value)
displays kernel and average shifted histograms categorized by firm (with an added category for the total), with all of the graphs in a single frame and the category value used as labels.
length.distplot cdf across(firm, dispname) within(income, bintype=quant, bincount=4)
constructs a categorical cdf graph with FIRM defining the across dimension, and INCOME defining the within dimension. Observations will be classified in the within dimension using the quartiles of INCOME.
Cross-references
For a description of distribution graphs, see
“Analytical Graph Types”.
See
“Graphing Data” for a detailed discussion of graphs in EViews, and
“Templates” for a discussion of graph templates.
See
Graph::graph for graph declaration and other graph types
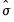 as the measure of dispersion), “iqr” (normal reference rule based on the interquartile range), “silverman” (normal reference rule with Silverman’s robust measure of dispersion), “freedman” (Freedman-Diaconis), “user” (user-specifed).
as the measure of dispersion), “iqr” (normal reference rule based on the interquartile range), “silverman” (normal reference rule with Silverman’s robust measure of dispersion), “freedman” (Freedman-Diaconis), “user” (user-specifed).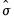 as the measure of dispersion.
as the measure of dispersion.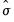 -normal reference rule, a kernel density plot, and a plot of the theoretical normal distribution fitted to the data. (Note that the “scale=dens” option in the
-normal reference rule, a kernel density plot, and a plot of the theoretical normal distribution fitted to the data. (Note that the “scale=dens” option in the